R for reproducible scientific analysis
Multi-Table Joins
Learning objectives
- Focus on the third tidy data principle
- Each variable forms a column.
- Each observation forms a row.
- Each type of observational unit forms a table.
- By able to use
dplyr’s join functions to merge tables
Joins
The third tidy data maxim states that each observation type gets its own table. The idea of multiple tables within a dataset will be familiar to anyone who has worked with a relational database but may seem foreign to those who have not.
The idea is this: Suppose we conduct a behavioral experiment that puts individuals in groups, and we measure both individual- and group-level variables. We should have a table for the individual-level variables and a separate table for the group-level variables. Then, should we need to merge them, we can do so using the join functions of dplyr.
The join functions are nicely illustrated in RStudio’s Data wrangling cheatsheet. Each function takes two data.frames and, optionally, the name(s) of columns on which to match. If no column names are provided, the functions match on all shared column names.
The different join functions control what happens to rows that exist in one table but not the other.
left_joinkeeps all the entries that are present in the left (first) table and excludes any that are only in the right table.right_joinkeeps all the entries that are present in the right table and excludes any that are only in the left table.inner_joinkeeps only the entries that are present in both tables.inner_joinis the only function that guarantees you won’t generate any missing entries.full_joinkeeps all of the entries in both tables, regardless of whether or not they appear in the other table.
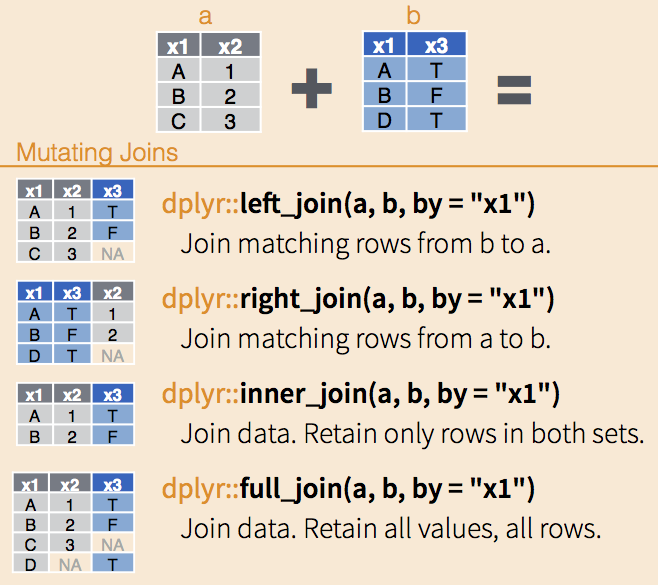
dplyr joins, via RStudio
We will practice on our continents data.frame from module 2 and the gapminder data.frame. Note how these are tidy data: We have observations at the level of continent and at the level of country, so they go in different tables. The continent column in the gapminder data.frame allows us to link them now. If continents data.frame isn’t in your Environment, load it and recall what it consists of:
load('data/continents.RDA')
continents continent area_km2 population percent_total_pop
1 Africa 30370000 1022234000 15.0
2 Americas 42330000 934611000 14.0
3 Antarctica 13720000 4490 0.0
4 Asia 43820000 4164252000 60.0
5 Europe 10180000 738199000 11.0
6 Oceania 9008500 29127000 0.4
We can join the two data.frames using any of the dplyr functions. We will pass the results to str to avoid printing more than we can read, and to get more high-level information on the resulting data.frames.
left_join(gapminder, continents) # A tibble: 1,704 × 9
country year pop continent lifeExp gdpPercap area_km2
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.801 779.4453 43820000
2 Afghanistan 1957 9240934 Asia 30.332 820.8530 43820000
3 Afghanistan 1962 10267083 Asia 31.997 853.1007 43820000
4 Afghanistan 1967 11537966 Asia 34.020 836.1971 43820000
5 Afghanistan 1972 13079460 Asia 36.088 739.9811 43820000
6 Afghanistan 1977 14880372 Asia 38.438 786.1134 43820000
7 Afghanistan 1982 12881816 Asia 39.854 978.0114 43820000
8 Afghanistan 1987 13867957 Asia 40.822 852.3959 43820000
9 Afghanistan 1992 16317921 Asia 41.674 649.3414 43820000
10 Afghanistan 1997 22227415 Asia 41.763 635.3414 43820000
# ... with 1,694 more rows, and 2 more variables: population <dbl>,
# percent_total_pop <dbl>
right_join(gapminder, continents)# A tibble: 1,705 × 9
country year pop continent lifeExp gdpPercap area_km2 population
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
1 Algeria 1952 9279525 Africa 43.077 2449.008 30370000 1022234000
2 Algeria 1957 10270856 Africa 45.685 3013.976 30370000 1022234000
3 Algeria 1962 11000948 Africa 48.303 2550.817 30370000 1022234000
4 Algeria 1967 12760499 Africa 51.407 3246.992 30370000 1022234000
5 Algeria 1972 14760787 Africa 54.518 4182.664 30370000 1022234000
6 Algeria 1977 17152804 Africa 58.014 4910.417 30370000 1022234000
7 Algeria 1982 20033753 Africa 61.368 5745.160 30370000 1022234000
8 Algeria 1987 23254956 Africa 65.799 5681.359 30370000 1022234000
9 Algeria 1992 26298373 Africa 67.744 5023.217 30370000 1022234000
10 Algeria 1997 29072015 Africa 69.152 4797.295 30370000 1022234000
# ... with 1,695 more rows, and 1 more variables: percent_total_pop <dbl>
These operations produce slightly different results, either 1704 or 1705 observations. Can you figure out why? Antarctica contains no countries so doesn’t appear in the gapminder data.frame. When we use left_join it gets filtered from the results, but when we use right_join it appears, with missing values for all of the country-level variables:
right_join(gapminder, continents) %>%
filter(continent == "Antarctica")# A tibble: 1 × 9
country year pop continent lifeExp gdpPercap area_km2 population
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
1 <NA> NA NA Antarctica NA NA 13720000 4490
# ... with 1 more variables: percent_total_pop <dbl>
There’s another problem in this data.frame – it has two population measures, one by continent and one by country and it’s not clear which is which! Let’s rename a couple of columns.
right_join(gapminder, continents) %>%
rename(country_pop = pop, continent_pop = population)# A tibble: 1,705 × 9
country year country_pop continent lifeExp gdpPercap area_km2
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
1 Algeria 1952 9279525 Africa 43.077 2449.008 30370000
2 Algeria 1957 10270856 Africa 45.685 3013.976 30370000
3 Algeria 1962 11000948 Africa 48.303 2550.817 30370000
4 Algeria 1967 12760499 Africa 51.407 3246.992 30370000
5 Algeria 1972 14760787 Africa 54.518 4182.664 30370000
6 Algeria 1977 17152804 Africa 58.014 4910.417 30370000
7 Algeria 1982 20033753 Africa 61.368 5745.160 30370000
8 Algeria 1987 23254956 Africa 65.799 5681.359 30370000
9 Algeria 1992 26298373 Africa 67.744 5023.217 30370000
10 Algeria 1997 29072015 Africa 69.152 4797.295 30370000
# ... with 1,695 more rows, and 2 more variables: continent_pop <dbl>,
# percent_total_pop <dbl>
Challenge – Putting the pieces together
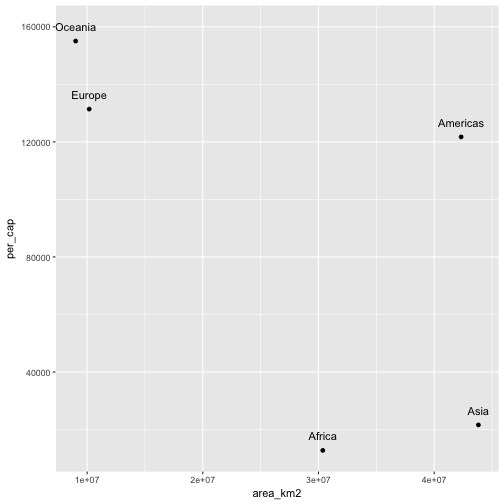
A colleague suggests that the more land area an individual has, the greater their gdp will be and that this relationship will be observable at any scale of observation. You chuckle and mutter “Not at the continental scale,” but your colleague insists. Test your colleague’s hypothesis by:
- Calculating the total GDP of each continent,
- Hint: Use
dplyr’sgroup_byandsummarize
- Hint: Use
- Joining the resulting data.frame to the
continentsdata.frame, - Calculating the per-capita GDP for each continent, and
- Plotting per-capita gdp versus population density.
Challenge solutions
Solution to Challenge – Putting the pieces together
library(ggplot2)
# Calculate country-level GDP
mutate(gapminder, GDP = gdpPercap * pop) %>%
# Group by continent
group_by(continent) %>%
# Calculate continent-level GDP
summarize(cont_gdp = sum(GDP)) %>%
# Join the continent-GDP data.frame to the continents data.frame
left_join(continents) %>%
# Calculate continent-level per-capita GDP
mutate(per_cap = cont_gdp / population) %>%
# Plot gdp versus land area
ggplot(aes(x = area_km2, y = per_cap)) +
# Draw points
geom_point() +
# And label them
geom_text(aes(label = continent), nudge_y = 5e3)